研究方向
1)AI底层技术:通过在模型中注入知识/规则,提升分子性质预测模型的泛化能力;
2)智能化学合成:利用深度学习、专家规则、量子化学构建化学合成智能体;
3)智能生物合成:利用AI设计、优化萜类合成酶和P450酶,辅助类药分子合成;
4)蛋白词表技术:从大模型中提取具有功能的蛋白语义单元(蛋白质的“官能团”),辅助药物、酶的设计。
实验室主页:https://www.tianlab-tsinghua.com/
学术贡献
田博学实验室近5年在化学信息学与AI制药领域取得系统性的研究成果,以通讯作者(含共同)身份发表系列研究论文,具体总结如下:
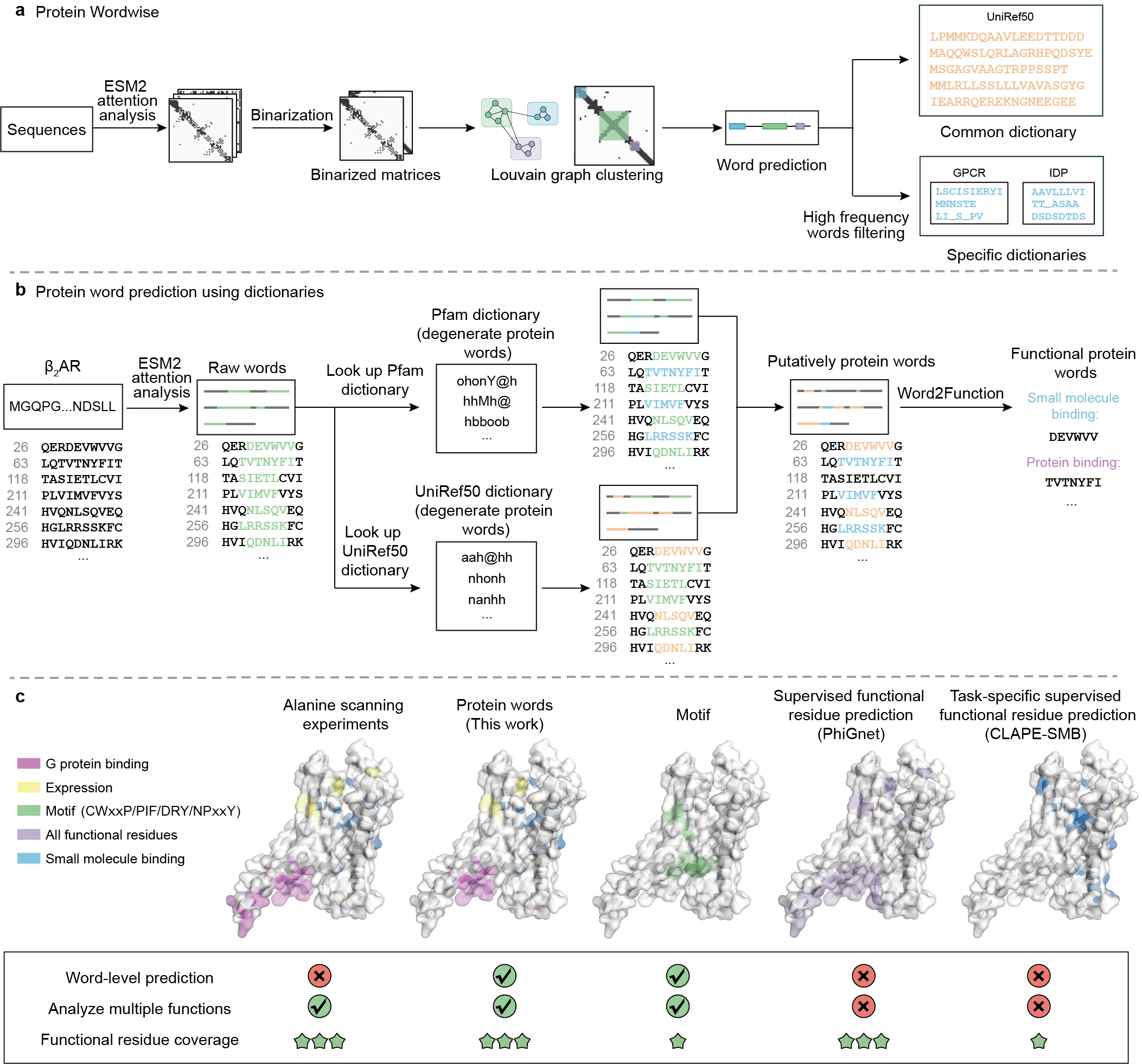
1)发展了蛋白词表技术,利用无监督学习分析大模型的注意力矩阵,解析蛋白语义单元,并利用监督学习映射到功能,有望成为下一代蛋白模拟、蛋白设计、药物设计、细胞模拟技术的基础 (B.Tian* BioRxiv 2025)。
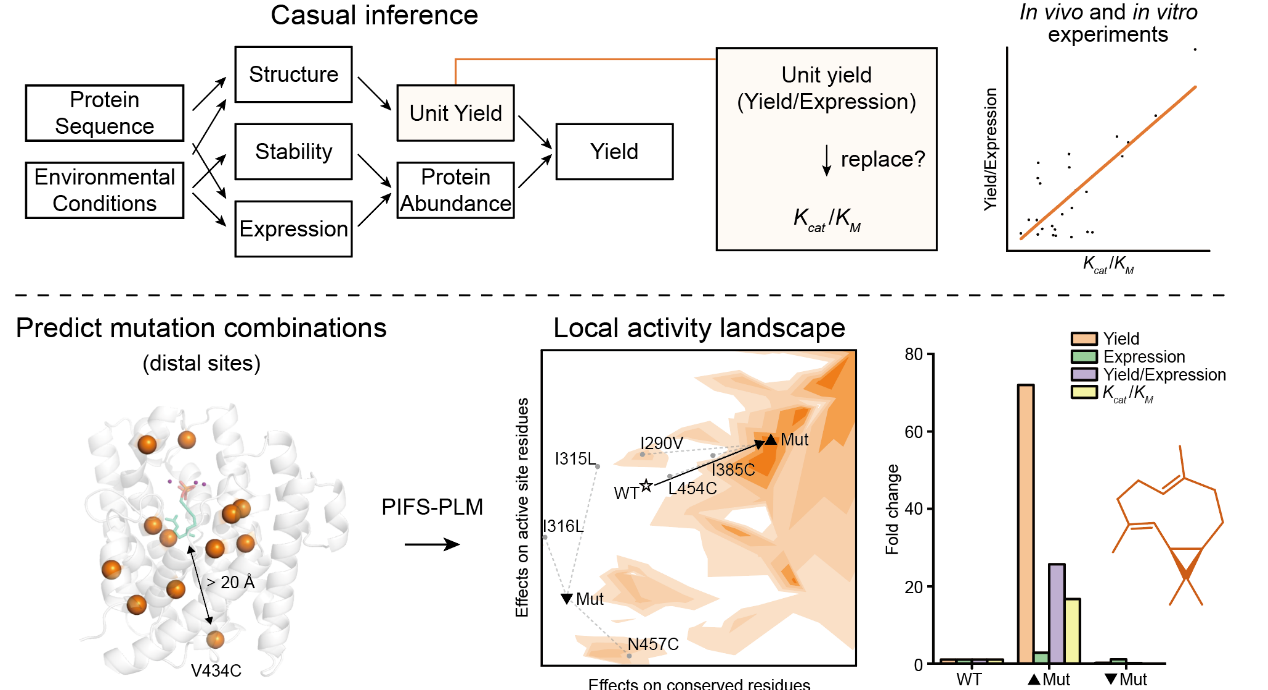
2)发展了物理启发式蛋白语言模型,利用远端突变提升了萜类合成酶的产物产量(70倍以上),辅助类药分子合成 (J. Qiao* B.Tian* Research Square 2024)。
3)发展了基于蛋白质语言模型的蛋白-DNA、蛋白-小分子的结合位点预测方法(B.Tian* Breif. Bioinform. 2024,B.Tian* J. Cheminform. 2024),抗体CDR结构预测 (F. Qian*, B.Tian* eLife 2024)、P450底物、产物预测方法 (B.Tian* J. Chem. Inf. Model. 2024,B.Tian* J. Chem. Inf. Model. 2025),辅助药物发现。
4)揭示了深度学习方法在分子性质预测中泛化能力不足的问题,训练获得了在MoleculeNet和MolData数据集上当前最佳模型(B.Tian* J. Chem. Inf. Model. 2025, B.Tian* J. Chem. Theory Comput. 2024),为化学信息学和AI制药领域提供了新思路。
5)在膜蛋白生物学机制研究方面,开展了转运蛋白(B.Tian, Y. Yuan*, C. Yan* Nature 2024)、离子通道(B.Tian*, X.Li*, B. Xiao* Neuron 2025)等体系的计算模拟。
6)利用量子化学方法研究了复杂环化、偶联、重排反应机制(B.Tian*, W.Deng* ACS Catal. 2021,W. Yang*,B.Tian*, W.Deng* ACS Catal. 2021, B.Tian*, X.Liao* Cell Rep. Phys. Sci. 2023, B.Tian*, Y.Tang* Org. Chem. Front. 2022)。
近5年论文发表情况:
1. Automatically Defining Protein Words for Diverse Functional Predictions Based on Attention Analysis of a Protein Language Model. Hedi Chen, Jingrui Zhong, Xiaochun Zhang, Jingke Chen, Lin Guo, Xiaoliang Xiong, Xiaonan Zhang, Xiangyu Liu, Bailong Xiao, Boxue Tian* bioRxiv 2025, https://doi.org/10.1101/2025.01.20.633699
2. GLMCyp: A Deep Learning-Based Method for CYP450-Mediated Reaction Site Prediction. Xuhai Huang, Jiamin Chang, Boxue Tian* Journal of Chemical Information and Modeling 2025, 65(5):2322-2335
3. SynthMol: A Drug Safety Prediction Framework Integrating Graph Attention and Molecular Descriptors into Pre-Trained Geometric Models. Zidong Su, Rong Zhang, Xiaoyu Fan, Boxue Tian* Journal of Chemical Information and Modeling 2025, 65:2256−2267
4. An intermediate open structure reveals the gating transition of the mechanically activated PIEZO1 channel. Sijia Liu ,Xuzhong Yang, Xudong Chen, Xiaochun Zhang, Jinghui Jiang, Jingyi Yuan, Wenhao Liu, Li Wang, Heng Zhou, Kun Wu, Boxue Tian*, Xueming Li*, Bailong Xiao* Neuron 2025,113,1-15
5. Enhancing Enzyme Activity with Mutation Combinations Guided by Few-shot Learning and Causal Inference. Research Square 2024, https://www.researchsquare.com/article/rs-5354708/v1
6. Protein–DNA binding sites prediction based on pre-trained protein language model and contrastive learning. Yufan Liu, Boxue Tian* Briefings in Bioinformatics 2024, 25(1):1-12
7. DeepP450: Predicting human P450 activities of small molecules by integrating pretrained protein language model and molecular representation. Jiamin Chang, Xiaoyu Fan, Boxue Tian* Journal of Chemical Information and Modeling 2024, 64(18):3149-3160
8. PointGAT: A quantum chemical property prediction model integrating graph attention and 3D geometry. Rong Zhang, Rongqing Yuan, Boxue Tian* Journal of Chemical Theory and Computation 2024, 20(10):4115-4128
9. Accurate prediction of CDR-H3 loop structures of antibodies with deep learning Hedi Chen,Xiaoyu Fan,Shuqian Zhu,Yuchan Pei,Xiaochun Zhang,Xiaonan Zhang,Lihang Liu,Feng Qian*,Boxue Tian* eLife 2024 12:RP91512
10. Molecular basis of human noradrenaline transporter reuptake and inhibition Jiaxin Tan,Yuan Xiao,Fang Kong,Xiaochun Zhang,Hanwen Xu,Angqi Zhu,Yiming Liu,Jianlin Lei,Boxue Tian,Yafei Yuan*,Chuangye Yan* Nature 2024, 632:921-929
11. Protein-small molecule binding site prediction based on a pre-trained protein language model with contrastive learning. Jue Wang, Yufan Liu & Boxue Tian* Journal of Cheminformatics 2024 16(1):125
12. Coupling of alkynes and aryl halides with nickel-catalyzed Sonogashira reactions. Hui Chen,Licheng Yao,Lin Guo,Yahu A. Liu,Boxue Tian,Xuebin Liao* Cell Reports Physical Science 2023, 5(1):102900
13. Elucidating the selectivity of dyotropic rearrangements of β-lactones: a computational survey. Jingyang Zhang, Yumiao Ma, Ke Qiu, Bo Li, Zhengwen Xue, Boxue Tian*,Yefeng Tang* Organic Chemistry Frontiers 2022, 9:329-341
14. Nickel-catalyzed enantioselective domino Heck/Sonogashira coupling for construction of C(sp)-C(sp3) bond-substituted quaternary carbon centers. Hui Chen, Zhenkang Ai, Lingmei Guo,Licheng Yao, Yaopeng Li, Buming Gu, Yixuan Zhang, Yahu A. Liu, Boxue Tian*, Xuebin Liao* Tetrahedron Chem 2022, 2:100021
15. Cooperative N-heterocyclic Carbene and Iridium Catalysis Enables Stereoselective and Regiodivergent [3 + 2] and [3 + 3] Annulation Reactions. Jian Zhang, Yanshan Gao, Buming Gu, Wulin Yang*, Boxue Tian*,Weiping Deng* ACS Catalysis 2021 11(7):3810-3821
16. Diastereo- and Enantioselective Synthesis of Eight-Membered Heterocycles via an Allylation/Ring Expansion Sequence Enabled by Multiple Catalysis. Wulin Yang*,Yuanlin Wang, Wen Li, Buming Gu, Siwen Wang, Xiaoyan Luo, Boxue Tian*, Weiping Deng* ACS Catalysis 2021 11(20):12557-12564

